Recent advancements in technologies and our growing knowledge of gene regulation have provided an exciting new direction to target leukemias and other cancers. These discoveries indicate that modifying a cell’s transcriptome by targeting a specific transcription factor (TF) can advantageously affect the the transcriptome of cancer cells. Based on this, we are leveraging our knowledge on SALL4 in cancer biology, gene regulation and structure to target the TF spalt-like 4 (SALL4) in cancer.
The lab is interested in using SALL4 as a paradigm to explore targeting transcription factors and downstream targets as therapeutic approaches in cancer.
Ongoing projects explore interaction between SALL4 and epigenetic regulators and SALL4’s roles in cancer metabolism for therapeutic targeting.
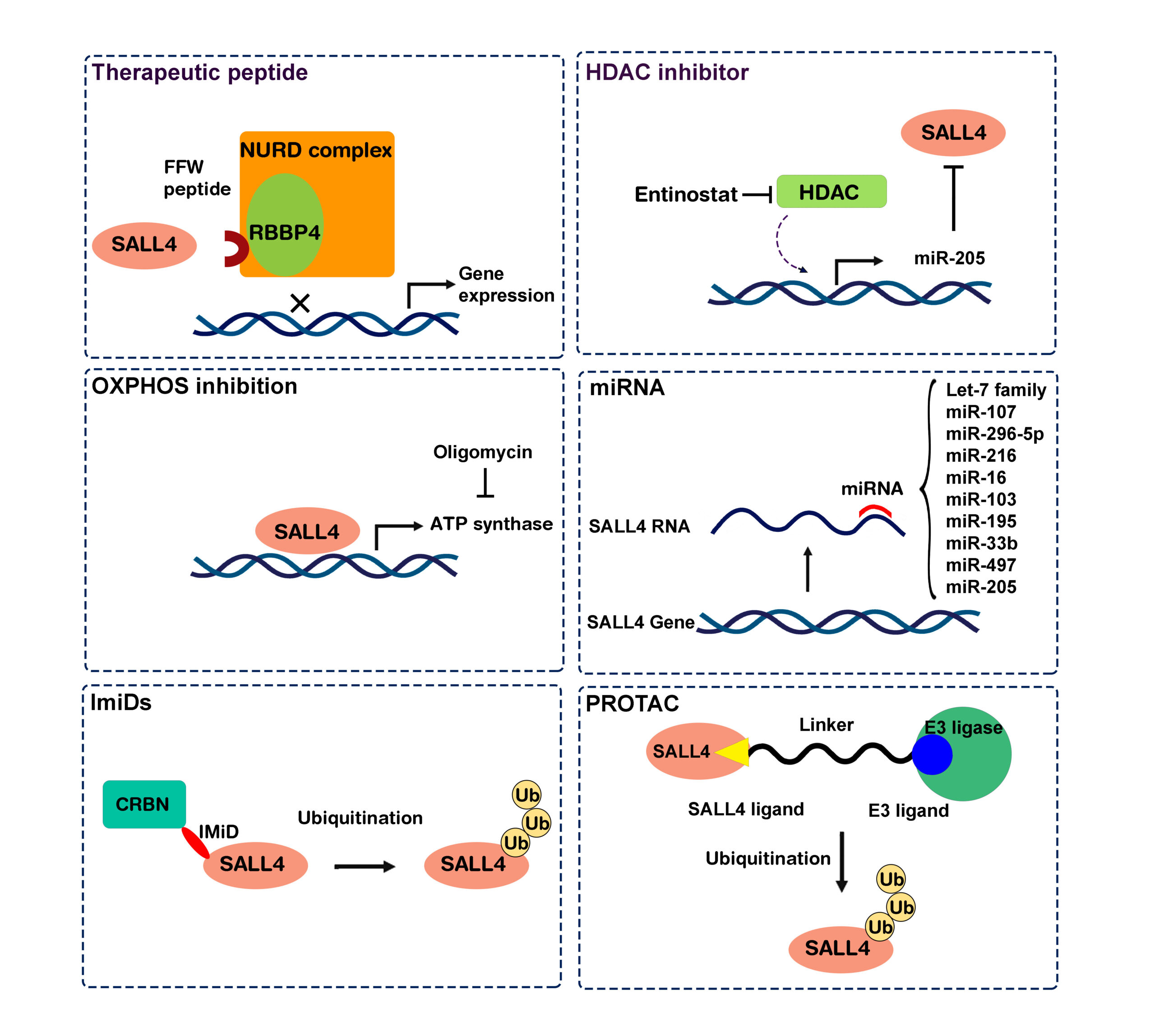
Scheme of the different drugs suggested to target SALL4 in cancer.
Moein et al. “SALL4: An Intriguing Therapeutic Target in Cancer Treatment”, Cells 2022.
The Chai lab is spearheading multiple approaches for SALL4 degradation and targeting in pre-clinical models.
SALL4 can function as a transcriptional repressor and activator through its interactions with both gene-repressing and activating epigenetic complexes. More specifically, SALL4 represses the expression of its target genes such as Phosphatase and Tensin homolog (PTEN) through its interaction with the Nucleosome Remodeling and Deacetylase complex (NuRD), a Histone Deacetylase (HDAC)-containing epigenetic repressor complex. SALL4 can also activate its target genes such as HOXA9 through its interaction with Mixed-Lineage Leukemia (MLL) protein, which is a key member of an epigenetic activator complex. We have further demonstrated that by blocking the interaction between SALL4 and NuRD with a peptide, we can inhibit its repression function. In addition to Pten and HoxA9, c-Myc is found to be a SALL4 target.
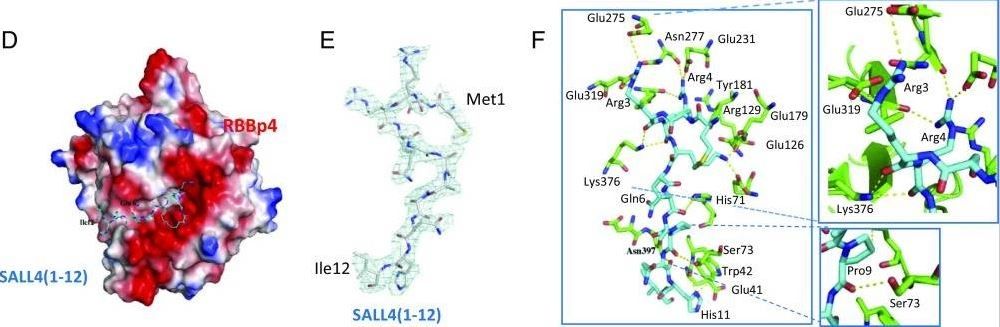
Structure of RBBp4–SALL4(1–12) complex and their binding affinity.
Liu BH et al. “Targeting cancer addiction for SALL4 by shifting its transcriptome with a pharmacologic peptide”, Proc Natl Acad Sci U S A, 2018
SALL4: a potentially actionable biomarker that guides combination therapy and precision medicine
- Cancer cells with high SALL4 expression exhibited more dependency on SALL4 compared with those with low SALL4 expression.
- A recent study developed a novel strategy to induce a cancer vulnerability by engineering a partial dependency upon SALL4 in lung cancer, hepatocellular carcinoma, and leukemic cell lines.
- Treatment of SALL4-negative cells with HMA (hypomethylating agents) resulted in transient upregulation of SALL4, which sensitized SALL4 negative cancer cells to Entinostat.
- HMA is widely used in myelodysplastic syndrome (MDS). SALL4 was essential for the efficiency of sequential treatment of HMA and Entinostat, suggesting a possible benefit of combination therapy for MDS.
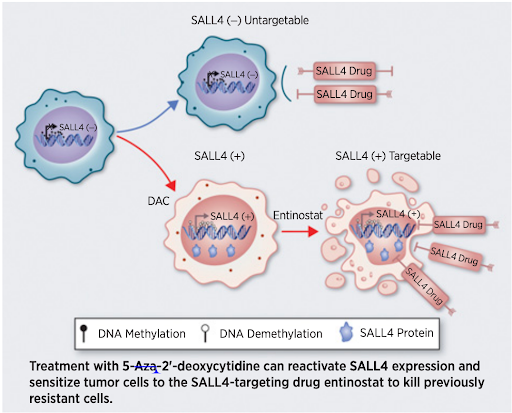
Cancer cells with high SALL4 expression exhibited more dependency on SALL4 compared with those with low SALL4 expression, which confers vulnerability to Entinostat.
Yang et al. “Targeting an Inducible SALL4-Mediated Cancer Vulnerability with Sequential Therapy” Cancer Research, 2021.