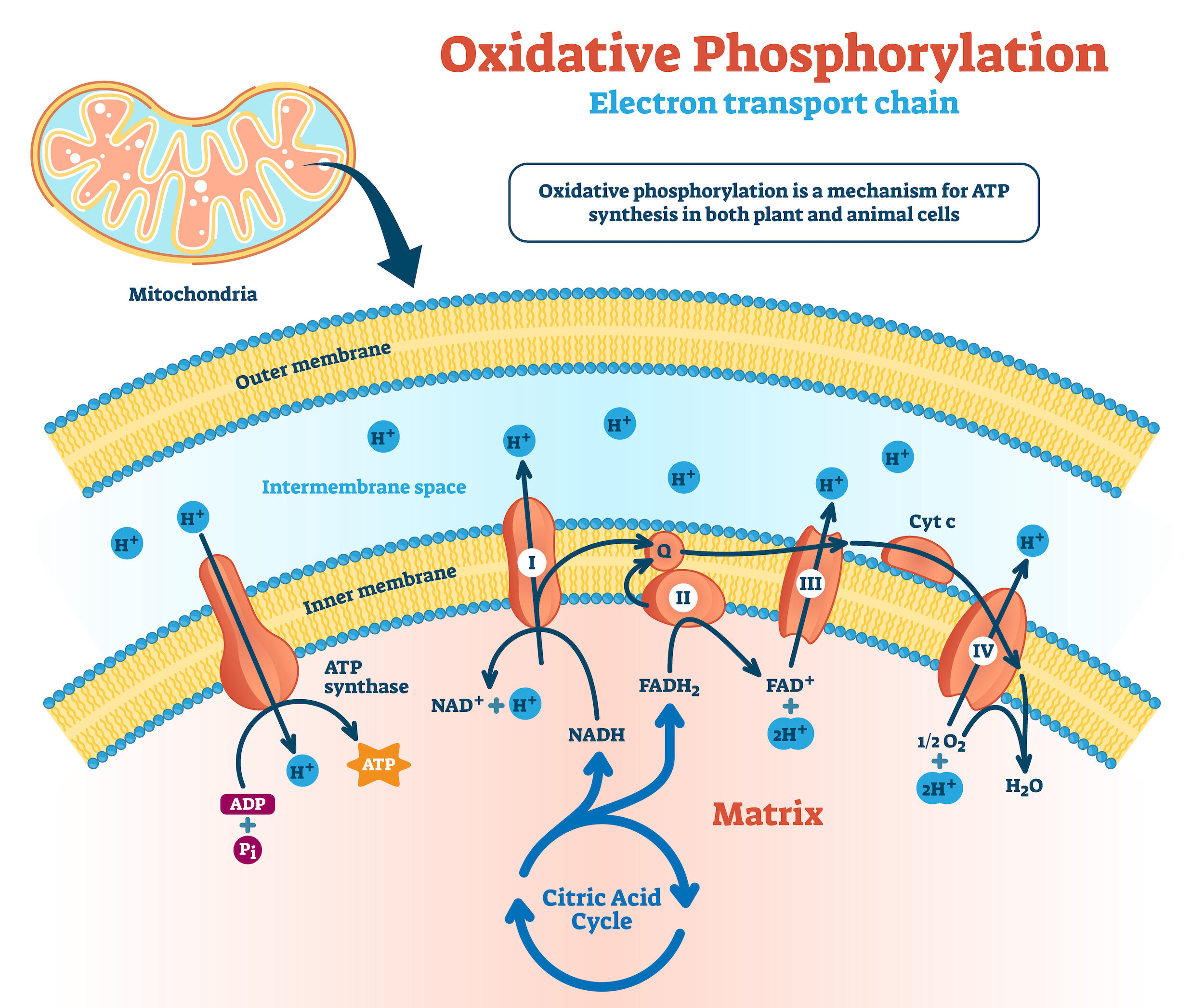
Mechanism(s) of SALL4: downstream targets and its role in metabolism
We have studied SALL4 downstream targets by performing genomic studies such as ChIP-Seq, Release using nuclease (CUT&RUN), RAN-seq et al. In doing so, we have identified that in addition to Pten, HoxA9, c-Myc, SALL4 targets include CBL-B, Bmi-1 and histone 3 lysine 9-specific demethylases (KDMs). We have also reported a novel metabolic function of SALL4 in upregulating mitochondrial oxidative phosphorylation in cancer. This upregulation results in an oxidative phosphorylation dependency in SALL4-expressing cancers that can be therapeutically targeted to suppress tumorigenesis.SALL4 diagnostics can benefit many cancer types where SALL4 is a targetable oncogene.